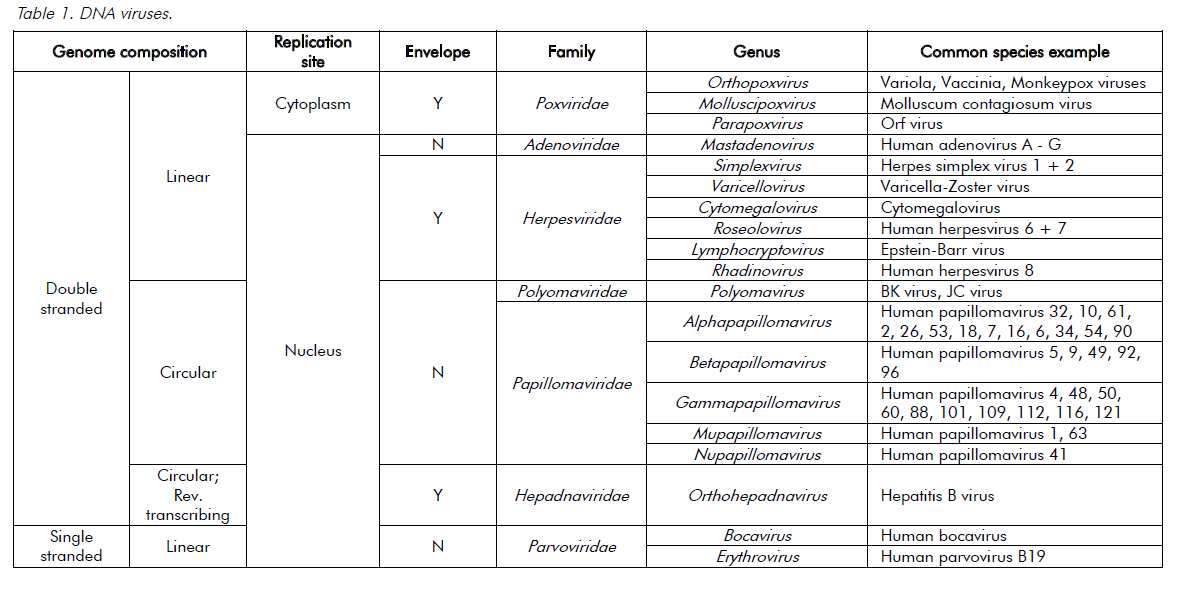
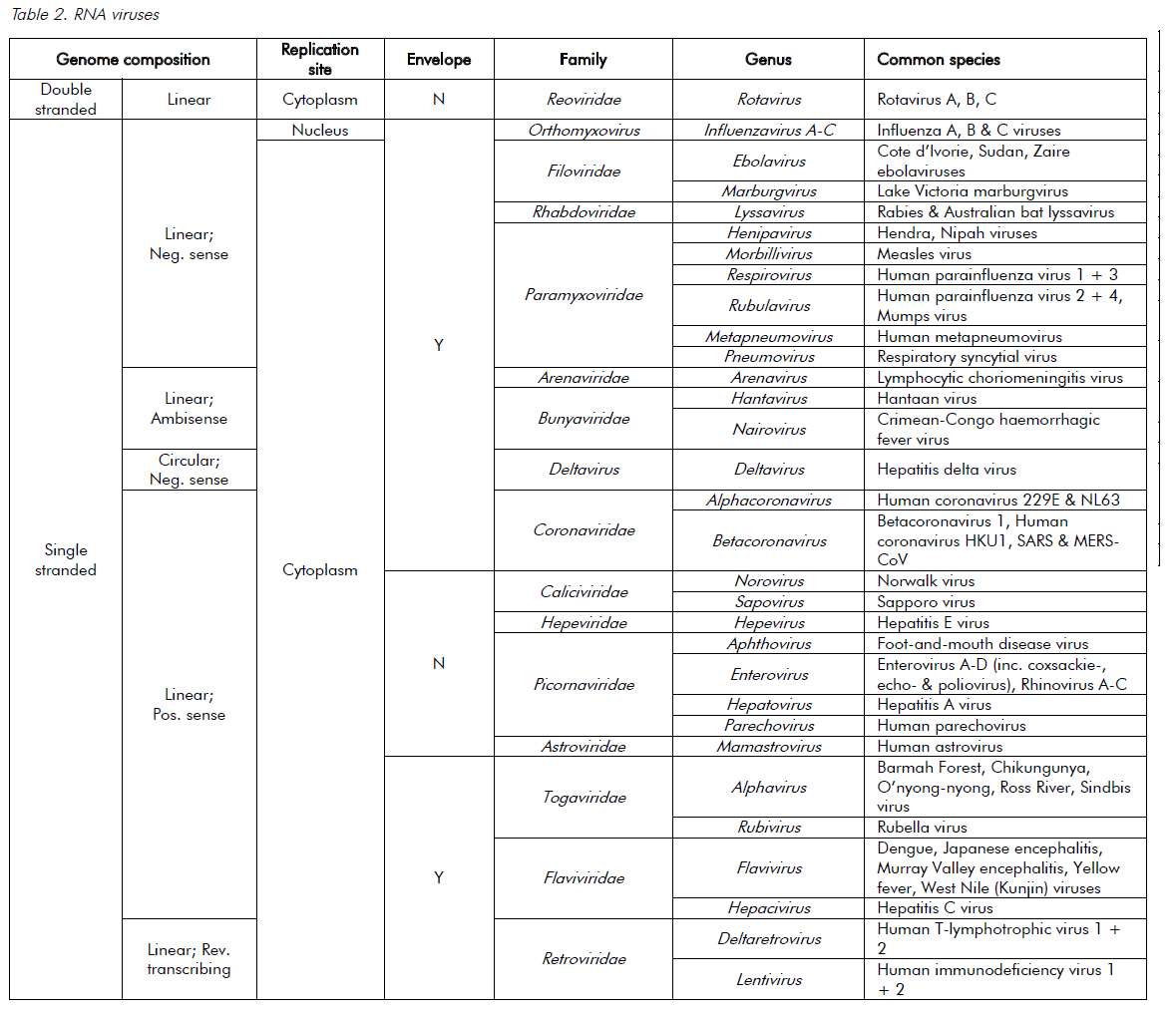
Viruses are ultramicroscopic, metabolically inert infectious agents that replicate only within living cells. Viruses occur universally and affect every animal, plant and eukaryotic micro-organism on the planet and, although some viruses affect the health of their hosts, most have very little or no impact. Unlike every other domain of life, which has double-stranded DNA (dsDNA) as its genetic program, virus genomes may be composed of either DNA or RNA. The genetic material is then arranged as either double or single strands. Single-stranded RNA (ssRNA) viruses are then further classified as being either positive-sense or negative-sense with respect to the messenger-RNA coding strand. The virus genome can have a linear or circular arrangement and consist of a single or multiple segments.
Compared with bacterial genomes, viral genomes are small, but can range over 100-fold in size (in other words, from 3000 nucleotides to 1 200 000 base pairs). Viral genomes are housed within a protein structure that forms the virus particle (see Figure 1). In some viruses, this nucleoprotein is surrounded by further protein or a lipid bilayer, referred to as the envelope. The outermost proteins of the virus particle allow the virus to recognise the correct host cells and gain entry into its cytoplasm. The simple composition of the virus structure means that it requires enzymes and other mechanisms from within the host cell in order to replicate. The survival of viruses is therefore dependant on the host species. A simplified classification of common DNA and RNA viruses affecting humans is presented in Tables 1 and 2.
Box 1. Diagnostic virology methods
- Cell culture
- Cytology
- Histopathology
- Electron microscopy
- Antigen detection
- Nucleic acid detection
- Serologic assays
Diagnostic virology
Laboratory techniques have advanced greatly in recent times, mainly with the advent of molecular methods. Different diagnostic laboratory methods are outlined in Box 1. Cultivation of viruses in cell culture can be extremely sensitive. One viable virion may be sufficient to initiate a positive result. Culture also has the potential to detect unsuspected, multiple or even novel viruses. Some of the shortcomings of this traditional method include: technical difficulty, expense, slow turnaround time, the requirement for different cell-lines for different viruses and that a viable virus is required for successful culture. For these reasons, viral culture has been replaced as a first-line diagnostic technique by simpler, faster methods.
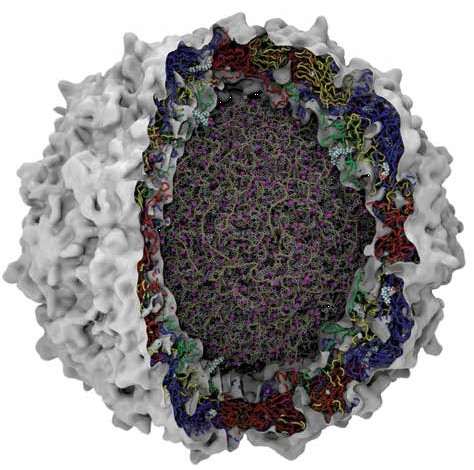
Figure 1. Schematic representation of a picornavirus virion. Cutaway representation of a complete picornavirus (poliovirus) virion. Image shows the genomic RNA in the core with associated Magnesium ions (purple). The protein chains that comprise the surrounding capsid are shown as Blue (VP1), Red (VP2), Yellow (VP3) and Green (VP4). Courtesy of Jason Roberts, National Enterovirus Reference Laboratory, Victorian Infectious Diseases Reference Laboratory, Melbourne, Australia.
Cytology can provide important clues to the presence of viral infection, and generally implicates a group of viruses rather than a specific virus. Cytological examination can be performed on smears, on slides prepared by cytocentrifugation of fluids and on ‘touch preps’. Tzank and Papanicolaou smears are examples of such techniques, which demonstrate the presence of HSV or VZV infection, or evidence of HPV infection, respectively. These are largely of historical interest only.
Histopathological findings suggestive of viral infection include intranuclear and intracytoplasmic inclusions, multinucleated giant cells and syncytia. Histopathology is enhanced by the use of immunohistochemistry to detect specific viral antigens and in-situ hybridisation to detect specific viral nucleic acids. This enhances the sensitivity and allows for identification of specific agents.
Electron microscopy (EM) provides a means to directly visualise the infecting virus and, like cell culture, allows for the identification of unsuspected, multiple or novel viruses. Many viruses have distinctive appearances under EM, which allows for identification to the family level, but this requires a skilled operator and dedicated and expensive equipment. Use of immunological techniques, such as immunogold labelling, can increase sensitivity somewhat and add a specific antigenic identification component to EM.
Antigen detection directly from clinical specimens can provide rapid information (within minutes to hours of specimen receipt) and is not dependent upon the presence of viable virus. The techniques most widely used are agglutination techniques, such as latex agglutination, fluorescent antibody (FA) staining, immunoperoxidase (IP) staining and enzyme immunoassay (EIA). These techniques are based on antibodies that specifically bind to the virus being sought. Although a versatile method, antigen detection is not as sensitive or as specific as nucleic acid amplification techniques and cannot be applied to all viruses, especially when the target virus has multiple serotypes and antigen variation.
Molecular methods have transformed the field of diagnostic virology. Viral nucleic acids, which are relatively stable to environmental conditions (DNA more so than RNA), can be detected regardless of the viability of the virus. Depending on the design of the primers and probes, amplification techniques, such as polymerase chain reaction (PCR), can either be very specific to a target (for example, the vaccine strain of measles virus) or target a genus or family for broader based detection (such as panflavivirus detection). This technology is now standard in most laboratories.
Commercial assays continue to simplify that process and are potentially able to identify multiple different targets from the one specimen. Furthermore, assessment of viral load allows for the determination of disease burden and response to therapy. Sequencing the amplified nucleic acid product also allows for virus identification and further characterisation such as subtyping, assessment of genetic drift, relatedness between strains and the assessment of drug-resistance mutations (for example, HIV genotype).
Serology is the method of measuring an antibody response to an infection and remains a powerful tool in viral diagnostics. There are many different types of assays, although the most common are EIA (or similar) assays. EIAs are now commonly incorporated into automated systems. Neutralisation and Western blot assays, however, remain the gold standard in many instances. Serologic diagnosis is dependent on the kinetics of the antibody response to infection, with an initial IgM phase and then subsequent IgG. Placental transfer of IgG antibodies limits the use of IgG-based serological testing in the newborn up to the age of 12–18 months. Depending on the infecting virus, the relation between the clinical syndrome, the viral and antibody kinetics can be markedly variable. Serology may be unhelpful in the acute setting, requiring a convalescent bleed for diagnosis. Serological evidence of an acute infection can be made based on IgG seroconversion, where the initial bleed was negative and the convalescent bleed positive.
A rising titre of a virus- specific antibody across two time periods can also suggest recent infection. The presence of virus-specific IgM antibodies in a single acute phase specimen can provide a rapid diagnosis in the correct clinical setting, but care should be taken given the possibility of false-positive results, either owing to cross-reacting antibodies or non-specific reactivity. The strength of IgG binding, known as IgG avidity, can also be used to assess the timing of an infection, where a low IgG avidity indicates a more recent infection (for example, CMV IgG avidity testing). Serology is also used for determining immunity (for example, hepatitis B virus, rubella virus and so forth).
Specimen collection and transport
The majority of errors occur in the pre- analytical phase of viral diagnostics. Beyond accurate labelling and safe and timely transport of the specimen, mistakes are often made either in the wrong test being ordered, an incorrectly collected specimen or a test requested at the wrong time relative to the clinical illness. Close liaison with the clinical microbiologist or infectious diseases service can often curtail these issues. Accurate and informative clinical notes will also enable the laboratory to best process, analyse and report the result accordingly.
Blood (whole blood, plasma or serum), urine, faeces, respiratory specimens, cerebrospinal fluid, biopsy tissue, ocularspecimens, vesicles and other skin lesions, and amniotic fluid should be considered for diagnostic testing. PCR techniques can also be applied retrospectively to histopathology specimens, where paraffin- embedded tissue is finely shaved and processed prior to nucleic acid extraction. The presence of PCR inhibitors may, however, limit the diagnostic yield.
Diagnosis of viral infections with a short viraemic phase (for example, flavivirus infections) may rely on serology rather than molecular testing, depending on the timing of symptoms. In the setting of HIV infection, serology remains the standard testing algorithm. HIV RNA viral load assays, which are not licenced in Australia as diagnostic tests, however, may be used as supplementary tests along with p24 antigen during the early phase of infection. In some clinical settings, a request for HIV pro-viral DNA is required and requires the collection of whole blood rather than plasma, to enable the collection and testing of the buffy coat (in other words, white blood cells).
Box 2. Common viral causes of different clinical syndromes
- Central nervous system infection* HSV, Enteroviruses, VZV, HHV6, arthropod-borne viruses, Influenza, HIV, LCM, Rabies
- Respiratory tract infections#Rhinovirus, Coronavirus, Adenovirus, Influenza virus, Parainfluenza virus, RSV, Human metapneumovirus, EBV
- Gastrointestinal infections Rotavirus, Norovirus, Astroviruses, enteric Adenoviruses
- Skin and mucous membrane infections HSV, VZV, enteroviruses, MCV, Orf virus, Papillomavirus
- Genital tract infections
HSV, Papillomavirus, MCV
- Viral hepatitis
Hepatitis A, B, C, D and E viruses, CMV, EBV, HIV, arthropod-borne viruses
- Cardiac infections†
Enterovirus, Adenovirus, CMV, Influenza virus, Parainfluenza virus, Mumps virus, Parvovirus B19
- Urinary tract infections
BK virus, Adenovirus, Hantavirus
- Ocular infections
Adenovirus, Enterovirus, HSV, VZV, CMV, Influenza virus, Mumps virus, Measles virus
- Childhood rash viral infections
Measles virus (First disease or Rubeola), Rubella virus (Third disease or German Mealses), Parvovirus B19 (Fifth disease or Erythema infectiosum), HHV6/7 (Sixth disease or Roseola infantum), VZV (chicken pox), Enterovirus
- Congenital infections
CMV, VZV, HSV, Parvovirus B19, LCM virus, HIV, Rubella virus
- Viral infections in immunocompromised patients CMV, HSV, VZV, EBV (causing PTLD), HHV6, HHV8 (causing KS), Adenovirus, BK virus (causing haemorrhagic cystitis), JC virus
(causing PML), Parvovirus B19, RSV, Enterovirus
- Viral haemorrhagic fevers
Ebola virus, Marburg virus, Lassa fever virus, Yellow fever virus, Dengue virus, Hantavirus, CCHF virus
- Blood-borne viruses
HIV, HTLV, Hepatitis B virus, Hepatitis C virus
*Includes meningitis and encephalitis; # includes upper and lower respiratory tract; † includes myocarditis and pericarditis.
HSV, Herpes simplex virus; VZV, Varicella-Zoster virus; HHV6/7, Human Herpes virus 6 or 7; HIV, Human immunodeficiency virus; RSV, Respiratory Syncytial virus; EBV, Epstein-Barr virus; MCV, Molluscum contagiosum virus; CMV, Cytomegalovirus; LCM, Lymphocytic Choriomeningitis virus; PTLD, post transplant lymphoproliferative disorder; KS, Kaposi’s sarcoma; PML, progressive multifocal leukoencephalopathy; CCHF, Crimean-Congo haemorrhagic fever virus; HTLV, Human T-lymphotrophic virus.
Correct sampling is vitally important for the sensitivity of any test. Flocked swabs, for example, can capture the greatest amount of infected material for analysis. When a nose and throat swab is collected, the posterior aspect of the nasopharynx should be sampled, rather than anterior nasal cavity. Furthermore, when a lower respiratory tract infection is strongly suspected and an initial nose and throat swab is negative, then assessment of a deep respiratory sample (for example, broncheoalveolar lavage) should be considered before ruling out the diagnosis. Interestingly, some viruses – for example, measles virus – will continue to be shed in the urine for longer than from the blood or nasopharynx.
The timing of molecular testing for the investigation of in-utero infections is especially important, given sampling the amniotic fluid either too early in gestation or too soon after maternal exposure could result in a false negative result. In suspected CMV infection, for example, sampling of the amniotic fluid should wait until later than six weeks after primary maternal infection and ≥21 weeks gestation for optimal sensitivity.
Box 3. Important viruses for obstetrics and gynaecology
- Human immunodeficiency virus
- Influenza viruses
- Parvovirus B19
- Rubella virus
- Cytomegalovirus
- Enterovirus
- Hepatitis B
- Hepatitis C
- Human papillomavirus
- Herpes simplex virus
Conclusions
Viruses can cause a wide variety of clinical presentations. Box 2 outlines the common viruses implicated in different clinical syndromes. The patient’s vaccination history, predisposing immunocompromising conditions or medications, geographic location, seasonality and age are all contributing factors. Reactivation of latent infections may also contribute to morbidity and mortality, although this may be actively prevented by screening and providing appropriate prophylaxis (such as lamivudine or entecavir therapy to prevent hepatitis B reactivation). Box 3 outlines the common viruses encountered in the obstetrics and gynaecology setting. It should be noted that a positive diagnostic test does not necessarily infer clinical disease, especially in the setting of viruses that cause latent infection (for example, Herpesviridae), or when molecular detection represents dead virus, rather than viable disease-causing virus. Similarly, the inability to detect the virus does not rule out the diagnosis. The correct predisposing conditions, the current clinical presentation and the investigation results all need to be weighed up against each other.
Further reading
Storch GA. Essentials of diagnostic virology. New York: Churchill Livingstone; 2000.
Dimmock NJ, Easton AJ, Leppard K. Introduction to modern virology. 6th ed. Malden, MA: Blackwell Pub.; 2007.
Versalovic J, American Society for Microbiology. Manual of clinical microbiology. 10th ed.
Washington, DC: ASM Press; 2011.
Knipe DM, Howley PM. Fields virology. 6th ed. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2013.
Palasanthiran P, Starr M, Jones C, Giles M. Management of Perinatal Infections. 3rd ed. Sydney, NSW: Australasian Society for Infectious Diseases Inc; 2014 (www.asid.net.au/documents/item/368, last accessed 23 March 2015).






Leave a Reply